I co-founded Precision Analytics with Erika, also holding a PhD from McGill and am an accredited statistician. While overseeing our …
A primer on bioinformatics
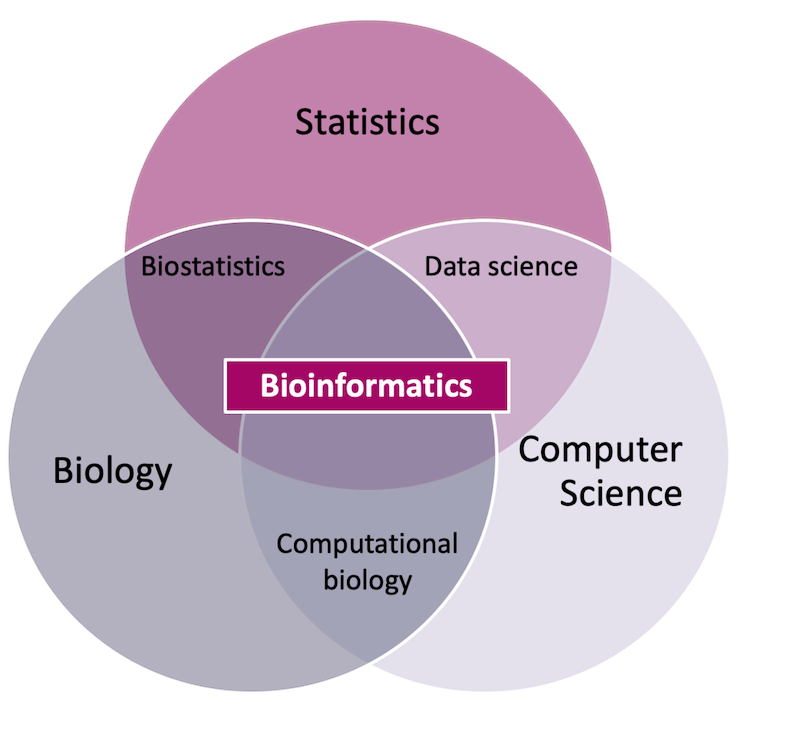
Bioinformatics has many definitions, but at its core it is an inherently applied discipline that uses statistics and computer science to study biological phenomena.
At Precision Analytics, we apply our training in biostatistics, mathematics, chemistry, biology, epidemiology and computer science to analyze data from a variety of sources and answer different business and research questions from our clients. Example data sources include:
- Genetic data from traditional or high-throughput sequencing, such as Sanger or next generation sequencing (NGS)
- In vitro data from bioassays, such as ELISA
- In vivo efficacy and pharmacokinetic studies in rats, mice, and other model organisms
On the computational side, methods such as string matching are important in the pre-processing of genetic data and looking for patterns. Because modern genomics involves large datasets, addressing computational challenges is key. For example, algorithms have been optimized to be computationally efficient to perform routine tasks like multiple sequence alignment .
The field of biostatistics introduces probabilistic analysis of data, such as drawing inferences from samples, estimating uncertainty, and predicting performance. These analyses could include:
- Descriptive statistics which are useful for understanding the distribution of data, standardizing data for comparisons, and assessing quality
- Hypothesis testing which is useful for comparing groups in a variety of applications; for example, to determine if a given gene is over-expressed in a set of tumour samples, or to test if people diagnosed with a disease have a higher frequency of a certain SNP
- Linear models such as variations of the classic ANOVA , to understand variation within and between groups
- More sophisticated statistical and mathematical models that can be used to better understand complex systems, calibrate or adjust for measurement errors, and account for uncertainty. For example, using hierarchical Bayesian models to account for autocorrelation in time course microarray data and extract biologically meaningful signals1
In the era of big data, machine learning and artificial intelligence present exciting opportunities in bioinformatics including for pattern recognition, correlation studies, and prediction.
Some particular statistical and computational challenges arise in the analysis of biological phenomena that require careful consideration. One such issue is of overtesting: because there are often a large number of comparisons, such as in genome-wide association studies, the probability of a false positive can become inflated2.
Adjusting for this overtesting is an ongoing research area in biostatistics and there are several interesting approaches currently used3. The best approach depends on the volume of testing, the computational complexity of the tests, and the research objective of the study.
For tips on performing your own bioinformatics analyses, see Open-source bioinformatics tools .
Find out more about our R training courses by contacting us at contact@precision-analytics.ca !
Ohlebusch, Enno. “Bioinformatics Algorithms.” Enno Ohlebusch (2013).
Sung, Wing-Kin. Algorithms in bioinformatics: A practical introduction. CRC Press, 2009.
Hensman, James, Neil D. Lawrence, and Magnus Rattray. “Hierarchical Bayesian modelling of gene expression time series across irregularly sampled replicates and clusters.” BMC bioinformatics 14.1 (2013): 1-12. ↩︎
Hayes, Ben. “Overview of statistical methods for genome-wide association studies (GWAS).” Genome-wide association studies and genomic prediction (2013): 149-169. ↩︎
Wei, Zhi, et al. “Multiple testing in genome-wide association studies via hidden Markov models.” Bioinformatics 25.21 (2009): 2802-2808. ↩︎
